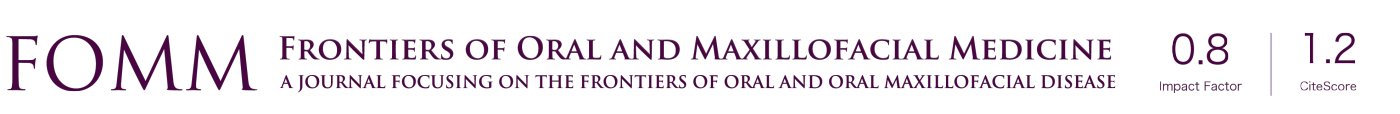Genetic factors underlying basal cell carcinoma risk: a narrative review
Introduction
Basal cell carcinoma (BCC) is the most prevalent form of malignant skin cancer with a good prognosis, originating from the basal layer of the epidermis and its appendages (1-4). Some clinical features, e.g., rodent ulcers, telangiectasias, pigmented and erosion areas, can be considered a characteristic of this disease (5,6). The incidence rates of BCC worldwide increase continuously owing to an aging population and widespread sun exposure. Still, the incidence estimation is imprecise since few population-based cancer registries and active nationwide surveillance systems exist in most countries and regions for monitoring and reporting the incidence of BCC (7,8). While BCC’s biological mechanism(s) is still unclear, environmental and genetic factors are primarily related to its pathogenesis (9,10). Some genetic agents have been considered the risk factors for BCC, e.g., Patched 1 (PTCH1) genes. In most cases, keratinocyte transformation occurs when the function of multiple genes (proto-oncogenes, tumor suppressor genes, and essential housekeeping genes) affected by mutations, causing hyperactivation of the HH protein family, leading to cell cycle deregulation, implicated as drivers in BCC formation (11,12). Moreover, the TP53 tumor suppressor gene is also commonly involved in the pathogenesis of BCC (3).
Therefore, this review provides an overview of BCC’s genetic pathogenesis. In this review, the PubMed database was searched with a search algorithm [(basal cell carcinoma) OR (BCC)] AND [(gene) OR (pathway)] to filter out relevant publications. Relevant researches omitted from this search algorithm were also selected from the specific full-text papers reference lists. We present the following article in accordance with the Narrative Review reporting checklist (available at https://fomm.amegroups.com/article/view/10.21037/fomm-21-70/rc).
Methods
The PubMed database was searched with a search algorithm [(basal cell carcinoma) OR (BCC)] AND [(gene) OR (pathway)], till May 2021, to filter out relevant publications. Relevant researches omitted from this search algorithm were also selected from the specific full-text papers reference lists. No language restrictions included in our search (Table 1).
Table 1
| Items | Specification |
|---|---|
| Date of search | 04 May 2021 |
| Databases and other sources searched | PubMed |
| Search terms used | [(basal cell carcinoma) OR (BCC)] AND [(gene) OR (pathway)] |
| Timeframe | Till 04 May 2021 |
| Inclusion and exclusion criteria | No searching restrictions |
| Selection process | All search and selection were done by Sitong Ju independently |
Hedgehog (HH) signaling pathway
The HH signaling pathway, i.e., the Hedgehog-Patched-Smoothened (Hh-Ptch-Smo), is an evolutionarily conserved pathway of signal transmission from the cell membrane to the nucleus, plays a vital role in the normal embryonic development of vertebrates (13) (Figure 1). Generally, the HH pathway is only active during embryonic development. A secreted protein, sonic HH ligand, binds to the transmembrane regulator receptor protein PTCH1 and inactivates it (14). Thus, the pathway is primarily inactive or poorly active in the adult organism. However, it can be activated in somatic and pluripotent stem cells in specific circumstances, such as tissue repair (15-17).
Malfunction or unusual activation of the pathway is associated with dysplasia and carcinogenesis (18-20). It behaves differently in different stages of various tumors, such as Gorlin syndrome (GS) (also known as Nevoid Basal Cell Carcinoma Syndrome, NBCCS), sporadic BCC, and medulloblastomas (21), among others. Gene variations (ligand-independent signaling) or HH signaling molecules overexpression (ligand-dependent signaling—autocrine or paracrine) can lead to aberrant activation of the pathway (22).
There are three proteins—HH ligand, PTCH, and Smoothened (SMO)—that participate in the activation of HH signaling (23).
The HH signaling pathway could be activated by SMO-activating variants or inactivating variants of PTCH1 or SUFU (HH signaling negative regulator), leading to constitutive activation of HH signaling without ligand (24). This connection was first found in GS patients with a PTCH1 mutation on chromosome 9 (14). According to the HH model, SMO repression is relieved following mutational inactivation of PTCH1 (25). Sonic HH (SHH), which is the strongest pathway activator, bind to PTCH1 and remove it from primary cilium and result in stimulation of SMO, triggers the activation of the transcription factors Gli1 (glioma-associated oncogene homolog) and/or Gli2 (26-30), resulting in cell proliferation. It has been involved in the development of BCC and other tumors, especially meningiomas and rhabdomyosarcomas (21).
Germline or somatic inactivating (loss-of-function) mutations of PTCH1, PTCH2, or SUFU, the activating (gain of function) mutations of SMO, or the amplification of GLI2 lead to the aberrant activation of HH (31), are responsible for over ninety percent of BCC cases in both sporadic BCC and GS. Mutations in the SUFU gene are the least common type have been seen in sporadic BCC (24) and specific hereditary syndromes, e.g., a subset of Gorlin BCC cases (5%) (32), multiple congenital infundibulocystic BCC syndrome cases (33), as well as some childhood-medulloblastoma families with “Gorlin-like” phenotype and hamartomatous skin lesions (34). Thus, there are three types of medulloblastoma caused by the aberrant activation of the HH signaling pathway: (I) NBCCS with SUFU mutations (20-fold increased risk than in the classic form) (32,35); (II) (a classic form of) NBCCS with PTCH1 mutations, and (III) NBCCS with PTCH2 mutations (a milder form than the previous one) (36). Incidentally, the PTCH2 gene mutation may also be the cause of sporadic BCC or sporadic medulloblastoma (37).
PTCH
PTCH is the receptor for HH protein, and two PTCH homologs have been isolated in vertebrates: PTCH1 and PTCH2 (38), both encode transmembrane receptors of the patched gene family in the SHH pathway and have diverse functions depending on their differential expression in epidermal development (26,39).
PTCH1 gene, primarily expressed in mesenchymal cells, is located on chromosome 9q22.3. It contains twenty-three exons and encodes a glycoprotein of 1,447 amino acids. PTCH1 binds to SHH proteins resulting in the stimulation of SMO, which acts as a transmembrane receptor of the HH signaling pathway and directs the embryonic growth of various organs in vertebrates (13,14,40). Its dysregulation is known to be essential in tumorigenesis, including BCC (41).
The most common PTCH1 mutations were frameshifts resulting in premature chain termination, which can cause premature termination of the PTCH protein (42-45). A recent survey found that PTCH1 had two mutated states—germinal and somatic mutation. More than 70% of patients with sporadic BCCs and xeroderma pigmentosum (XP) related BCCs detect somatic mutations of PTCH1, suggesting that abnormal activation of HH signaling is a precondition for both BCC associated with the GS and sporadic BCCs development (11,24,46-50).
Furthermore, a study shows that the PTCH1 expression level was higher in BCC with both mutation types than those with only germinal mutations (51). National Cancer Institute of USA evaluated 18 NBCCS families with whole-exome sequencing and revealed that eighty-nine percent of these families expressed disease-causing mutations in PTCH1 (52). Similarly, Undén et al. also identified that PTCH1 mRNA was over-expressed in BCC cells compared to the expression level in non-tumorous epidermal cells (53).
The PTCH2 gene is a protein-coding gene located on 1p34.1. It encodes a 1203-amino acid protein (transmembrane receptors) of the patched gene family in the SHH pathway, mainly expressed in the skin and testicular epithelial cells (54). Previously, a Chinese Han family with NBCCS carrying a heterozygous germline missense mutation in PTCH2 was reported by Fan et al. (55). And Fujii et al. reported a 13-year-old girl diagnosed with NBCCS based on multiple keratocystic odontogenic tumors and rib anomalies, who carried a frameshift mutation in the PTCH2 gene (c.1172_1173delCT) suggested that PTCH2 variants can also cause NBCCS, albeit with a milder phenotype (36). However, a healthy female with a PTCH2 homozygous frameshift variant was reported, and she did not have any of the symptoms included in the diagnostic criteria for NBCCS (56). Whether PTCH2 variants are associated with NBCCS is still unclear, as the number of cases with PTCH2 mutations remains limited. The accumulation of such patients is expected to clarify their characteristic phenotype further.
TP53
TP53 is the second essential gene in BCC carcinogenesis. It maintains genomic stability by encoding the P53 protein that regulates the cell cycle, induces apoptosis, and activates DNA repair. Mutations of TP53 have been identified in 20 to over 60 percent of sporadic BCCs (11). It is known that P53 is related to the early development of many different types of cancer, including BCC. And the loss of heterozygosity (LOH) of P53 appears mutually exclusive with PTCH1 (11). Furthermore, P53 protein is involved in the aging process of keratinocytes, and a function of the protein may facilitate BCCs growth in this case (11).
Ultraviolet (UV) radiation exposure as a leading environmental agent can result in several oncogenes and anti-oncogenes mutations implicated as drivers in BCC formation (4,11,57). The pattern of genetic mutations involved in BCC pathogenesis is consistent with DNA mutations caused by ultraviolet (UVA and UVB), owing to the “UV signature” mutations they harbored (58,59). Cyclobutane dimers and pyrimidine (6–4) pyrimidone photoproducts [6-4PPs, repaired through a process known as nucleotide excision repair (NER) after damage] (60) are used for UV landmark producing (C to T or CC to TT transversions) (61,62). Within these genes, the mutational profiles of BCCs reveal evidence of UV-induced mutagenesis. In most cases, mutations identified in both the HH/Patched/SMO pathway and TP53 are consistent with UV radiation-induced mutagenesis (63), which shows that repairing UV-induced DNA damage can reduce the carcinogenicity of BCC. In addition to UV-induced changes, other factors are also linked to mutations in BCCs, such as oxidative stress (11,46).
Besides BCC-specific mutational drivers (PTCH1, TP53), there are pigmentary-traits-determined genes relevant for germline polymorphisms, including melanocortin-1 receptor (MC1R), the human homolog of agouti signaling protein gene (ASIP), and tyrosinase (TYR). The increased risk of BCC development is associated with single nucleotide polymorphisms, in which ASIP and TYR genes are involved and responsible for the regulation of melanin hormones (64-66).
Personal and/or family history of skin cancer, coupled with fair complexion, light/red hair color, light eye color, and poor tanning ability (high sensitivity to UV exposure), are well-known BCC risk factors (7,67-69). Pigmentation is a multigenic trait, and MC1R, a membrane G coupled protein involved in melanin production, is a major factor in determining skin and hair color (70). Several studies indicate that mutations of the MC1R gene are significantly associated with BCC risk and exert carcinogenic pigmentation-independent effects (65,66,71). A family history of skin cancer is closely related to an increased risk of developing BCC under 40 (odds ratio 2.49, 95% CI: 1.80–3.45), independent of the MC1R phenotype (72).
Tyrosinase, encoded by TYR gene, is located on the human chromosome 11q14-q21 (73). Tyrosinase is a copper-containing enzyme and plays an important role in melanin production by catalyzing the oxidation of tyrosine to dopamine (DOPA), DOPA to L-dopaquinone, and 5,6-dihydroxyindole to 5,6-in-dolequinone in skin, hair, and eye (74). Mutations in the TYR gene may cause oculocutaneous albinism (OCA), a genetic disorder related to a higher risk of non-melanoma skin cancer (NMSC) (64).
Concerning multiple BCCs genetic susceptibilities, several studies have found an association between main factors, such as the number of BCCs, the vitamin D receptor, tumor necrosis factor, polymorphisms shown by the cytochrome P-450 (CYP2D6) and the glutathione S-transferase (GST) supergene family. The importance of these factors in cellular mechanisms such as metabolism and detoxification has been proved (75-78). However, the connection between these genetic polymorphisms, oncogenesis, and the clinical phenotype remains unclear.
Several pieces of research have identified genetic variants that may affect BCC risks (79-82). For example, a genome-wide association meta-analysis reveals that single nucleotide polymorphisms in genes participating in DNA excision repair may be involved in the etiopathogenesis of BCC (83).
Inherited disorders
Certain inherited disorders, including GS, XP, and Bazex-Dupré-Christol syndrome (BDCS), are considered genetic risk factors for BCC, predisposing BCC at an early age (84-86).
GS
GS is a rare multi-system disorder of autosomal dominant inheritance. In most cases, caused by germline inactivating mutations of the human PTCH1 on the chromosome 9q22.3–q31, infrequently by mutations in the SUFU gene and PTCH2. It is distinguished by dysplasia and postnatal tumors, including odontogenic keratocysts, medulloblastoma, and multiple BCCs with an average age of 20 to 21 (36,55,87-89). The incidence of GS ranges from 1:56,000 to 1:164,000 among the general population (84).
XP
XP is a rare autosomal recessive disorder. Mutations in genes involved in repairing UV-induced DNA damage are the primary pathogenesis of XP, and mutations in XPA, C, and V were detected in 75% of all patients (90). Clinical features include early-onset pigmented skin changes and the early development of skin cancers. According to a 39-year follow-up study of 106 XP patients conducted by the National Institutes of Health (NIH, USA), 69 (65%) patients with skin cancer have a higher risk of suffering from NMSC and melanoma than the general population. Moreover, the median age at first diagnosis of NMSC was significantly younger than melanoma (91).
Bazex-Dupré-Christol syndrome
This syndrome is an X-linked dominant disorder characterized by congenital hypotrichosis, follicular atrophoderma, and multiple BCCs (92). It may be accompanied by milia, ichthyosis, neurological symptoms, and visceral malignancies (93). According to the latest research, the BDCS gene is localized on the Xq25-27.1 and yet unspecified.
Rombo syndrome
In 1981, Michaëlsson et al. (88) described Rombo syndrome for the first time. An autosomal dominant inheritance across at least four generations was detected, leading to early-onset and frequent BCCs in the middle of the nineteen-thirties. This syndrome is characterized by dilation of the peripheral blood vessels with cyanosis and skin follicle atrophy features, which can be seen in the first decade of life. Dilation of the capillaries and milia-like papules on the face are particularly prominent in adults; eyelash and eyebrow loss or abnormalities, as well as less common capillary hemangiomas, were also observed. Histological analysis revealed regional loss of elastin clumps and elastin.
OCA
OCA is a group of autosomal recessive disorders. It manifests as a series of visual impairments and hypopigmentation of the skin and hair due to impaired melanin biosynthesis. There is an increased risk of early-onset skin cancer in those with OCA. OCA is caused by mutations in genes encoding proteins involved in the melanin biosynthesis pathway, which include melanogenic enzymes [i.e., TYR, tyrosinase-related protein 1 (TYRP1)] and specific transport proteins found in melanosomes (64). The TYR enzyme catalyzes the first step in melanin biosynthesis by oxidizing L-tyrosine to DOPA (dihydroxy-L-phenylalanine). In patients with OCA, squamous cell carcinoma is the most prevalent cancer type that arises; BCC and melanoma also occur (94). Malignant melanoma (MM) is considered rare in patients with OCA compared to NMSC (95).
Other genes
BRCA1 and BRCA2
BRCA1 and BRCA2 are tumor suppressor genes encoding proteins that help repair damaged DNA. Specific mutations of these genes may increase breast and ovarian cancer risks and also lead to several additional types of cancer.
BRCA1 mutations confer a predisposition to various cancer types, including melanomas, mesotheliomas, clear cell renal cancer, and BCCs (96-98). And a study has proved that BRCA2 can increase the risk of BCCs (99). Hence, more careful skin surveillance and photoprotection should be promoted for patients with the cancers mentioned above and/or known carriers of BRCA1 and BRCA2.
CTLA-4
CTLA-4, which can affect the immune response, may also influence BCC predisposition. CTLA-4 is a protein-coding gene that delivers suppressive signals to regulatory T cells and is associated with the immune tolerance induced by UV light. In a case-control study, genetic mutations of CTLA-4 affect the risk of developing BCC, particularly in those with a high frequency of severe tanning (100). In addition, CTLA-4 polymorphism (rs5742909) has been reported that may influence the susceptibility to multiple BCCs (101).
AS3MT
According to multiple studies, chronic arsenic exposure may cause superficial multicentric BCC (102-106) and the intake of arsenic mostly from contaminated drinking water, seafood, or medications. Some genetic factors related may affect BCC risks, such as AS3MT [arsenic (+3 oxidation state) methyltransferase gene, the major arsenic-metabolizing gene] mutation (107,108). AS3MT is a protein-coding gene encoding the arsenite methyltransferase enzyme and telomere length (105,109). A detailed study conducted by Srinivas et al. investigates the effect of telomere length on the disease risk. The result shows that compared with 533 healthy controls, the telomere length of 528 BCC-combined arsenic-exposed cases significantly decreased, which indicates that with arsenic exposure, decreased telomere length of individuals raises the risk of developing BCC. And there was a synergistic effect in those with the highest arsenic exposure and the shortest telomeres (109).
It should be noticed that beyond the genes mentioned above (Table 2), other tumor-related genes and pathways are also involved in the pathogenesis of BCC (11,46,63,94,121).
Table 2
| Gene | No. of samples analyzed | Mutations (%) | LOH (%) | Year | References |
|---|---|---|---|---|---|
| PTCH 1 | 37 | 32.4 | 24.3 | 1996 | (50) |
| 24 | 54.2 | Na | 1996 | (14) | |
| 55 | Na | 66.7 | 1996 | (110) | |
| 26 | 11.8 | 38.2 | 1998 | (45) | |
| 15 | 40 | 53.3 | 2002 | (111) | |
| 14 | 64.3 | 92.8 | 2005 | (112) | |
| 12 | 75 | 41.7 | 2005 | (24) | |
| 12 | 8.3 | 40 | 2011 | (113) | |
| 31 | 54.8 | 43.5 | 2013 | (114) | |
| 42 | 66.7 | 52.6 | 2014 | (63) | |
| 293 | 73 | 55 | 2016 | (46) | |
| 20 | 60 | 10 | 2019 | (51) | |
| 191 | 58.6 | 23 | 2020 | (115) | |
| 14 | 50 | Na | 1992 | (116) | |
| 27 | 56 | Na | 1993 | (117) | |
| TP53 | 18 | 61.1 | 5.5 | 1996 | (110) |
| 24 | 45.8 | Na | 1996 | (14) | |
| 20 | 35 | Na | 1999 | (118) | |
| 15 | 33 | Na | 2002 | (111) | |
| 148 | 47.3 | Na | 2003 | (119) | |
| 42 | 40.5 | 7.9 | 2005 | (24) | |
| 30 | 20 | Na | 2014 | (120) | |
| 12 | 66.7 | Na | 2014 | (63) | |
| 293 | 61 | 17 | 2016 | (46) | |
| 191 | 31.4 | Na | 2020 | (115) | |
| SMO | 47 | 6.38 | Na | 1998 | (25) |
| 42 | 9.5 | Na | 2005 | (24) | |
| 293 | 20 | Na | 2016 | (46) | |
| SUFU | 42 | 2.4 | Na | 2005 | (24) |
| 293 | 8 | 5 | 2016 | (46) |
LOH, loss of heterozygosity; BCC, basal cell carcinoma; Na, not applicable.
A genetic analysis study of 293 BCCs concluded that gene mutations of eighty-five percent cases were related to the HH pathway (PTCH1 73%, or SMO 20%, and SUFU 8 %) and TP53 (61%) (46). Furthermore, additional driver mutations in other cancer-related genes, such as MYCN (30%), PTPN14 (23%), PPP6C (15%), STK19 (10%), LATS1 (8%), can also be observed in 85% of cases. The up-regulation of N-Myc and Hippo-YAP pathway target genes (MYCN, PTPN14, and LATS1) shows the potential relevance in BCC tumorigenesis and progression (46). These factors are likely to account for the enormous phenotypic and biological variation in BCC. In a study of 12 sporadic BCCs and normal skin, mutations were identified in several known or presumptive cancer genes (CSMD1, DPP10, NOTCH1, and PREX2) via whole-exome sequencing; meanwhile, mutational hotspots were detected in STAT5B, CRNKL1, and NEBL (63). The relevance of these mutations to the genesis of BCC, however, is still unclear. Recently, Sławińska et al. reported that STAT3 and IL‑6 polymorphism are associated with the risk of BCC (122).
Conclusions
The hereditary basis of BCCs can vary from targeted mutations in the HH signaling pathway to deficiencies of tumor suppressors and melanin synthesis. These may lead to DNA damage and promotes BCC growth. The knowledge and characterization of the BCC genetic factors could underlie the development of new therapies and ultimately reduce BCC’s worldwide burden.
Acknowledgments
Funding: This study was supported by the Fundamental Research Funds for the Central Universities (No. 2021FZZX005-15), and the State Scholarship Fund from China Scholarship Council (No. 202108080189).
Footnote
Provenance and Peer Review: This article was commissioned by the editorial office, Frontiers of Oral and Maxillofacial Medicine for the series “Diagnosis and Treatment of Periorbital Basal Cell Carcinoma”. The article has undergone external peer review.
Reporting Checklist: The authors have completed the Narrative Review reporting checklist. Available at https://fomm.amegroups.com/article/view/10.21037/fomm-21-70/rc
Peer Review File: Available at https://fomm.amegroups.com/article/view/10.21037/fomm-21-70/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://fomm.amegroups.com/article/view/10.21037/fomm-21-70/coif). The series “Diagnosis and Treatment of Periorbital Basal Cell Carcinoma” was commissioned by the editorial office without any funding or sponsorship. LMH and VK served as the unpaid Guest Editors of the special series. LMH serves as an unpaid editorial board member of Frontiers of Oral and Maxillofacial Medicine from September 2020 to August 2022. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Sreekantaswamy S, Endo J, Chen A, et al. Aging and the treatment of basal cell carcinoma. Clin Dermatol 2019;37:373-8. [Crossref] [PubMed]
- Peris K, Fargnoli MC, Garbe C, et al. Diagnosis and treatment of basal cell carcinoma: European consensus-based interdisciplinary guidelines. Eur J Cancer 2019;118:10-34. [Crossref] [PubMed]
- Pópulo H, Boaventura P, Vinagre J, et al. TERT promoter mutations in skin cancer: the effects of sun exposure and X-irradiation. J Invest Dermatol 2014;134:2251-7. [Crossref] [PubMed]
- Cameron MC, Lee E, Hibler BP, et al. Basal cell carcinoma: Epidemiology; pathophysiology; clinical and histological subtypes; and disease associations. J Am Acad Dermatol 2019;80:303-17. [Crossref] [PubMed]
- Marzuka AG, Book SE. Basal cell carcinoma: pathogenesis, epidemiology, clinical features, diagnosis, histopathology, and management. Yale J Biol Med 2015;88:167-79. [PubMed]
- Wozniak-Rito A, Zalaudek I, Rudnicka L. Dermoscopy of basal cell carcinoma. Clin Exp Dermatol 2018;43:241-7. [Crossref] [PubMed]
- Verkouteren JAC, Ramdas KHR, Wakkee M, et al. Epidemiology of basal cell carcinoma: scholarly review. Br J Dermatol 2017;177:359-72. [Crossref] [PubMed]
- Wu S, Han J, Li WQ, et al. Basal-cell carcinoma incidence and associated risk factors in U.S. women and men. Am J Epidemiol 2013;178:890-7. [Crossref] [PubMed]
- Oh ST, Lee J, Yang KJ, et al. Increased Immunoreactivity of LGR4 in Histologically Aggressive Basal Cell Carcinoma. Ann Dermatol 2018;30:630-3. [Crossref] [PubMed]
- Tilli CM, Van Steensel MA, Krekels GA, et al. Molecular aetiology and pathogenesis of basal cell carcinoma. Br J Dermatol 2005;152:1108-24. [Crossref] [PubMed]
- Pellegrini C, Maturo MG, Di Nardo L, et al. Understanding the Molecular Genetics of Basal Cell Carcinoma. Int J Mol Sci 2017;18:2485. [Crossref] [PubMed]
- Hussein MR. Ultraviolet radiation and skin cancer: molecular mechanisms. J Cutan Pathol 2005;32:191-205. [Crossref] [PubMed]
- Bale AE, Yu KP. The hedgehog pathway and basal cell carcinomas. Hum Mol Genet 2001;10:757-62. [Crossref] [PubMed]
- Johnson RL, Rothman AL, Xie J, et al. Human homolog of patched, a candidate gene for the basal cell nevus syndrome. Science 1996;272:1668-71. [Crossref] [PubMed]
- Lowry WE, Richter L, Yachechko R, et al. Generation of human induced pluripotent stem cells from dermal fibroblasts. Proc Natl Acad Sci U S A 2008;105:2883-8. [Crossref] [PubMed]
- Zhou JX, Jia LW, Liu WM, et al. Role of sonic hedgehog in maintaining a pool of proliferating stem cells in the human fetal epidermis. Hum Reprod 2006;21:1698-704. [Crossref] [PubMed]
- Le H, Kleinerman R, Lerman OZ, et al. Hedgehog signaling is essential for normal wound healing. Wound Repair Regen 2008;16:768-73. [Crossref] [PubMed]
- Kobasa D, Takada A, Shinya K, et al. Enhanced virulence of influenza A viruses with the haemagglutinin of the 1918 pandemic virus. Nature 2004;431:703-7. [Crossref] [PubMed]
- Bailey JM, Mohr AM, Hollingsworth MA. Sonic hedgehog paracrine signaling regulates metastasis and lymphangiogenesis in pancreatic cancer. Oncogene 2009;28:3513-25. [Crossref] [PubMed]
- Ma X, Sheng T, Zhang Y, et al. Hedgehog signaling is activated in subsets of esophageal cancers. Int J Cancer 2006;118:139-48. [Crossref] [PubMed]
- Hahn H, Wicking C, Zaphiropoulous PG, et al. Mutations of the human homolog of Drosophila patched in the nevoid basal cell carcinoma syndrome. Cell 1996;85:841-51. [Crossref] [PubMed]
- Rubin LL, de Sauvage FJ. Targeting the Hedgehog pathway in cancer. Nat Rev Drug Discov 2006;5:1026-33. [Crossref] [PubMed]
- Cross SS, Bury JP. The Hedgehog signalling pathways in human pathology. Current Diagnostic Pathology 2004;10:157-68. [Crossref]
- Reifenberger J, Wolter M, Knobbe CB, et al. Somatic mutations in the PTCH, SMOH, SUFUH and TP53 genes in sporadic basal cell carcinomas. Br J Dermatol 2005;152:43-51. [Crossref] [PubMed]
- Xie J, Murone M, Luoh SM, et al. Activating Smoothened mutations in sporadic basal-cell carcinoma. Nature 1998;391:90-2. [Crossref] [PubMed]
- Booth DR. The hedgehog signalling pathway and its role in basal cell carcinoma. Cancer Metastasis Rev 1999;18:261-84. [Crossref] [PubMed]
- Dahmane N, Lee J, Robins P, et al. Activation of the transcription factor Gli1 and the Sonic hedgehog signalling pathway in skin tumours. Nature 1997;389:876-81. [Crossref] [PubMed]
- Eichberger T, Regl G, Ikram MS, et al. FOXE1, a new transcriptional target of GLI2 is expressed in human epidermis and basal cell carcinoma. J Invest Dermatol 2004;122:1180-7. [Crossref] [PubMed]
- Wicking C, Smyth I, Bale A. The hedgehog signalling pathway in tumorigenesis and development. Oncogene 1999;18:7844-51. [Crossref] [PubMed]
- Skoda AM, Simovic D, Karin V, et al. The role of the Hedgehog signaling pathway in cancer: A comprehensive review. Bosn J Basic Med Sci 2018;18:8-20. [Crossref] [PubMed]
- Chung JH, Bunz F. A loss-of-function mutation in PTCH1 suggests a role for autocrine hedgehog signaling in colorectal tumorigenesis. Oncotarget 2013;4:2208-11. [Crossref] [PubMed]
- Smith MJ, Beetz C, Williams SG, et al. Germline mutations in SUFU cause Gorlin syndrome-associated childhood medulloblastoma and redefine the risk associated with PTCH1 mutations. J Clin Oncol 2014;32:4155-61. [Crossref] [PubMed]
- Schulman JM, Oh DH, Sanborn JZ, et al. Multiple Hereditary Infundibulocystic Basal Cell Carcinoma Syndrome Associated With a Germline SUFU Mutation. JAMA Dermatol 2016;152:323-7. [Crossref] [PubMed]
- Huq AJ, Walsh M, Rajagopalan B, et al. Mutations in SUFU and PTCH1 genes may cause different cutaneous cancer predisposition syndromes: similar, but not the same. Fam Cancer 2018;17:601-6. [Crossref] [PubMed]
- Taylor MD, Liu L, Raffel C, et al. Mutations in SUFU predispose to medulloblastoma. Nat Genet 2002;31:306-10. [Crossref] [PubMed]
- Fujii K, Ohashi H, Suzuki M, et al. Frameshift mutation in the PTCH2 gene can cause nevoid basal cell carcinoma syndrome. Fam Cancer 2013;12:611-4. [Crossref] [PubMed]
- Smyth I, Narang MA, Evans T, et al. Isolation and characterization of human patched 2 (PTCH2), a putative tumour suppressor gene inbasal cell carcinoma and medulloblastoma on chromosome 1p32. Hum Mol Genet 1999;8:291-7. [Crossref] [PubMed]
- Zaphiropoulos PG, Undén AB, Rahnama F, et al. PTCH2, a novel human patched gene, undergoing alternative splicing and up-regulated in basal cell carcinomas. Cancer Res 1999;59:787-92. [PubMed]
- Gailani MR, Bale AE. Developmental genes and cancer: role of patched in basal cell carcinoma of the skin. J Natl Cancer Inst 1997;89:1103-9. [Crossref] [PubMed]
- Hahn H, Christiansen J, Wicking C, et al. A mammalian patched homolog is expressed in target tissues of sonic hedgehog and maps to a region associated with developmental abnormalities. J Biol Chem 1996;271:12125-8. [Crossref] [PubMed]
- Stone DM, Hynes M, Armanini M, et al. The tumour-suppressor gene patched encodes a candidate receptor for Sonic hedgehog. Nature 1996;384:129-34. [Crossref] [PubMed]
- Zhang H, Ping XL, Lee PK, et al. Role of PTCH and p53 genes in early-onset basal cell carcinoma. Am J Pathol 2001;158:381-5. [Crossref] [PubMed]
- Lam CW, Leung CY, Lee KC, et al. Novel mutations in the PATCHED gene in basal cell nevus syndrome. Mol Genet Metab 2002;76:57-61. [Crossref] [PubMed]
- Wicking C, Shanley S, Smyth I, et al. Most germ-line mutations in the nevoid basal cell carcinoma syndrome lead to a premature termination of the PATCHED protein, and no genotype-phenotype correlations are evident. Am J Hum Genet 1997;60:21-6. [PubMed]
- Aszterbaum M, Rothman A, Johnson RL, et al. Identification of mutations in the human PATCHED gene in sporadic basal cell carcinomas and in patients with the basal cell nevus syndrome. J Invest Dermatol 1998;110:885-8. [Crossref] [PubMed]
- Bonilla X, Parmentier L, King B, et al. Genomic analysis identifies new drivers and progression pathways in skin basal cell carcinoma. Nat Genet 2016;48:398-406. [Crossref] [PubMed]
- Epstein EH. Basal cell carcinomas: attack of the hedgehog. Nat Rev Cancer 2008;8:743-54. [Crossref] [PubMed]
- D'Errico M, Calcagnile A, Canzona F, et al. UV mutation signature in tumor suppressor genes involved in skin carcinogenesis in xeroderma pigmentosum patients. Oncogene 2000;19:463-7. [Crossref] [PubMed]
- Bodak N, Queille S, Avril MF, et al. High levels of patched gene mutations in basal-cell carcinomas from patients with xeroderma pigmentosum. Proc Natl Acad Sci U S A 1999;96:5117-22. [Crossref] [PubMed]
- Gailani MR, Ståhle-Bäckdahl M, Leffell DJ, et al. The role of the human homologue of Drosophila patched in sporadic basal cell carcinomas. Nat Genet 1996;14:78-81. [Crossref] [PubMed]
- Martinez MF, Romano MV, Martinez AP, et al. Nevoid Basal Cell Carcinoma Syndrome: PTCH1 Mutation Profile and Expression of Genes Involved in the Hedgehog Pathway in Argentinian Patients. Cells 2019;8:144. [Crossref] [PubMed]
- Gianferante DM, Rotunno M, Dean M, et al. Whole-exome sequencing of nevoid basal cell carcinoma syndrome families and review of Human Gene Mutation Database PTCH1 mutation data. Mol Genet Genomic Med 2018;6:1168-80. [Crossref] [PubMed]
- Undén AB, Zaphiropoulos PG, Bruce K, et al. Human patched (PTCH) mRNA is overexpressed consistently in tumor cells of both familial and sporadic basal cell carcinoma. Cancer Res 1997;57:2336-40. [PubMed]
- Larsson NG, Wang J, Wilhelmsson H, et al. Mitochondrial transcription factor A is necessary for mtDNA maintenance and embryogenesis in mice. Nat Genet 1998;18:231-6. [Crossref] [PubMed]
- Fan Z, Li J, Du J, et al. A missense mutation in PTCH2 underlies dominantly inherited NBCCS in a Chinese family. J Med Genet 2008;45:303-8. [Crossref] [PubMed]
- Altaraihi M, Wadt K, Ek J, et al. A healthy individual with a homozygous PTCH2 frameshift variant: Are variants of PTCH2 associated with nevoid basal cell carcinoma syndrome? Hum Genome Var 2019;6:10. [Crossref] [PubMed]
- Zak-Prelich M, Narbutt J, Sysa-Jedrzejowska A. Environmental risk factors predisposing to the development of basal cell carcinoma. Dermatol Surg 2004;30:248-52. [PubMed]
- Ikehata H, Ono T. The mechanisms of UV mutagenesis. J Radiat Res 2011;52:115-25. [Crossref] [PubMed]
- Boeckmann L, Martens MC, Kakkassery V, et al. Molecular genetic investigations as the basis for targeted treatment of basal cell carcinoma of the eye. Ophthalmologe 2020;117:106-12. [Crossref] [PubMed]
- Sinha RP, Häder DP. UV-induced DNA damage and repair: a review. Photochem Photobiol Sci 2002;1:225-36. [Crossref] [PubMed]
- Yamamoto J, Plaza P, Brettel K. Repair of (6-4) Lesions in DNA by (6-4) Photolyase: 20 Years of Quest for the Photoreaction Mechanism. Photochem Photobiol 2017;93:51-66. [Crossref] [PubMed]
- Guo Y, Rokohl AC, Kopecky A, et al. Periocular basal cell carcinoma—current treatment concepts. Ann Eye Sci 2021;6:18. [Crossref]
- Jayaraman SS, Rayhan DJ, Hazany S, et al. Mutational landscape of basal cell carcinomas by whole-exome sequencing. J Invest Dermatol 2014;134:213-20. [Crossref] [PubMed]
- Gudbjartsson DF, Sulem P, Stacey SN, et al. ASIP and TYR pigmentation variants associate with cutaneous melanoma and basal cell carcinoma. Nat Genet 2008;40:886-91. [Crossref] [PubMed]
- Han J, Kraft P, Colditz GA, et al. Melanocortin 1 receptor variants and skin cancer risk. Int J Cancer 2006;119:1976-84. [Crossref] [PubMed]
- Box NF, Duffy DL, Irving RE, et al. Melanocortin-1 receptor genotype is a risk factor for basal and squamous cell carcinoma. J Invest Dermatol 2001;116:224-9. [Crossref] [PubMed]
- NCCN Clinical Practice Guidelines in Oncology. Basal Cell Skin Cancer. Version 1.2018. 2017.
- Trakatelli M, Morton C, Nagore E, et al. Update of the European guidelines for basal cell carcinoma management. Eur J Dermatol 2014;24:312-29. [Crossref] [PubMed]
- Rubin AI, Chen EH, Ratner D. Basal-cell carcinoma. N Engl J Med 2005;353:2262-9. [Crossref] [PubMed]
- From Genetics Home Reference. MC1R gene. Genetics home reference 2018. Available online: https://ghr.nlm.nih.gov/gene/MC1R. Accessed 15 Jan 2018.
- Bastiaens MT, ter Huurne JA, Kielich C, et al. Melanocortin-1 receptor gene variants determine the risk of nonmelanoma skin cancer independently of fair skin and red hair. Am J Hum Genet 2001;68:884-94. [Crossref] [PubMed]
- Berlin NL, Cartmel B, Leffell DJ, et al. Family history of skin cancer is associated with early-onset basal cell carcinoma independent of MC1R genotype. Cancer Epidemiol 2015;39:1078-83. [Crossref] [PubMed]
- Tomita Y, Takeda A, Okinaga S, et al. Human oculocutaneous albinism caused by single base insertion in the tyrosinase gene. Biochem Biophys Res Commun 1989;164:990-6. [Crossref] [PubMed]
- Körner A, Pawelek J. Mammalian tyrosinase catalyzes three reactions in the biosynthesis of melanin. Science 1982;217:1163-5. [Crossref] [PubMed]
- Ramachandran S, Fryer AA, Lovatt TJ, et al. Combined effects of gender, skin type and polymorphic genes on clinical phenotype: use of rate of increase in numbers of basal cell carcinomas as a model system. Cancer Lett 2003;189:175-81. [Crossref] [PubMed]
- Yengi L, Inskip A, Gilford J, et al. Polymorphism at the glutathione S-transferase locus GSTM3: interactions with cytochrome P450 and glutathione S-transferase genotypes as risk factors for multiple cutaneous basal cell carcinoma. Cancer Res 1996;56:1974-7. [PubMed]
- Hajeer AH, Lear JT, Ollier WE, et al. Preliminary evidence of an association of tumour necrosis factor microsatellites with increased risk of multiple basal cell carcinomas. Br J Dermatol 2000;142:441-5. [Crossref] [PubMed]
- Ramachandran S, Lear JT, Ramsay H, et al. Presentation with multiple cutaneous basal cell carcinomas: association of glutathione S-transferase and cytochrome P450 genotypes with clinical phenotype. Cancer Epidemiol Biomarkers Prev 1999;8:61-7. [PubMed]
- Stacey SN, Gudbjartsson DF, Sulem P, et al. Common variants on 1p36 and 1q42 are associated with cutaneous basal cell carcinoma but not with melanoma or pigmentation traits. Nat Genet 2008;40:1313-8. [Crossref] [PubMed]
- Stacey SN, Sulem P, Masson G, et al. New common variants affecting susceptibility to basal cell carcinoma. Nat Genet 2009;41:909-14. [Crossref] [PubMed]
- Rafnar T, Sulem P, Stacey SN, et al. Sequence variants at the TERT-CLPTM1L locus associate with many cancer types. Nat Genet 2009;41:221-7. [Crossref] [PubMed]
- Stacey SN, Sulem P, Jonasdottir A, et al. A germline variant in the TP53 polyadenylation signal confers cancer susceptibility. Nat Genet 2011;43:1098-103. [Crossref] [PubMed]
- Lin Y, Chahal HS, Wu W, et al. Association study of genetic variation in DNA repair pathway genes and risk of basal cell carcinoma. Int J Cancer 2017;141:952-7. [Crossref] [PubMed]
- Schierbeck J, Vestergaard T, Bygum A. Skin Cancer Associated Genodermatoses: A Literature Review. Acta Derm Venereol 2019;99:360-9. [Crossref] [PubMed]
- Jaju PD, Ransohoff KJ, Tang JY, et al. Familial skin cancer syndromes: Increased risk of nonmelanotic skin cancers and extracutaneous tumors. J Am Acad Dermatol 2016;74:437-51; quiz 452-4. [Crossref] [PubMed]
- Cameron MC, Lee E, Hibler BP, et al. Basal cell carcinoma: Contemporary approaches to diagnosis, treatment, and prevention. J Am Acad Dermatol 2019;80:321-39. [Crossref] [PubMed]
- Xu LL, Li TJ. PTCH2 gene alterations in keratocystic odontogenic tumors associated with nevoid basal cell carcinoma syndrome. Beijing Da Xue Xue Bao Yi Xue Ban 2008;40:15-8. [PubMed]
- Michaëlsson G, Olsson E, Westermark P. The Rombo syndrome: a familial disorder with vermiculate atrophoderma, milia, hypotrichosis, trichoepitheliomas, basal cell carcinomas and peripheral vasodilation with cyanosis. Acta Derm Venereol 1981;497-503. [PubMed]
- MacDonald DS. A systematic review of the literature of nevoid basal cell carcinoma syndrome affecting East Asians and North Europeans. Oral Surg Oral Med Oral Pathol Oral Radiol 2015;120:396-407. [Crossref] [PubMed]
- DiGiovanna JJ, Kraemer KH. Shining a light on xeroderma pigmentosum. J Invest Dermatol 2012;132:785-96. [Crossref] [PubMed]
- Bradford PT, Goldstein AM, Tamura D, et al. Cancer and neurologic degeneration in xeroderma pigmentosum: long term follow-up characterises the role of DNA repair. J Med Genet 2011;48:168-76. [Crossref] [PubMed]
- Torrelo A, Sprecher E, Mediero IG, et al. What syndrome is this? Bazex-Dupre-Christol syndrome. Pediatr Dermatol 2006;23:286-90. [Crossref] [PubMed]
- AlSabbagh MM, Baqi MA. Bazex-Dupré-Christol syndrome: review of clinical and molecular aspects. Int J Dermatol 2018;57:1102-6. [Crossref] [PubMed]
- Kiprono SK, Chaula BM, Beltraminelli H. Histological review of skin cancers in African Albinos: a 10-year retrospective review. BMC Cancer 2014;14:157. [Crossref] [PubMed]
- Ozaki S, Funasaka Y, Otsuka Y, et al. Melanotic Malignant Melanoma in Oculocutaneous Albinism Type 4. Acta Derm Venereol 2017;97:287-8. [Crossref] [PubMed]
- Wadt KA, Aoude LG, Johansson P, et al. A recurrent germline BAP1 mutation and extension of the BAP1 tumor predisposition spectrum to include basal cell carcinoma. Clin Genet 2015;88:267-72. [Crossref] [PubMed]
- de la Fouchardière A, Cabaret O, Savin L, et al. Germline BAP1 mutations predispose also to multiple basal cell carcinomas. Clin Genet 2015;88:273-7. [Crossref] [PubMed]
- Mochel MC, Piris A, Nose V, et al. Loss of BAP1 Expression in Basal Cell Carcinomas in Patients With Germline BAP1 Mutations. Am J Clin Pathol 2015;143:901-4. [Crossref] [PubMed]
- Ginsburg OM, Kim-Sing C, Foulkes WD, et al. BRCA1 and BRCA2 families and the risk of skin cancer. Fam Cancer 2010;9:489-93. [Crossref] [PubMed]
- Welsh MM, Applebaum KM, Spencer SK, et al. CTLA4 variants, UV-induced tolerance, and risk of non-melanoma skin cancer. Cancer Res 2009;69:6158-63. [Crossref] [PubMed]
- Sławińska M, Zabłotna M, Nowicki RJ, et al. FOXP3 and CTLA-4 genetic variants' influence on the susceptibility and clinical course of basal cell carcinoma. Postepy Dermatol Alergol 2021;38:455-60. [Crossref] [PubMed]
- Kim TH, Seo JW, Hong YS, et al. Case-control study of chronic low-level exposure of inorganic arsenic species and non-melanoma skin cancer. J Dermatol 2017;44:1374-9. [Crossref] [PubMed]
- Mayer JE, Goldman RH. Arsenic and skin cancer in the USA: the current evidence regarding arsenic-contaminated drinking water. Int J Dermatol 2016;55:e585-91. [Crossref] [PubMed]
- Cheng PS, Weng SF, Chiang CH, et al. Relationship between arsenic-containing drinking water and skin cancers in the arseniasis endemic areas in Taiwan. J Dermatol 2016;43:181-6. [Crossref] [PubMed]
- Karagas MR, Gossai A, Pierce B, et al. Drinking Water Arsenic Contamination, Skin Lesions, and Malignancies: A Systematic Review of the Global Evidence. Curr Environ Health Rep 2015;2:52-68. [Crossref] [PubMed]
- Boonchai W, Green A, Ng J, et al. Basal cell carcinoma in chronic arsenicism occurring in Queensland, Australia, after ingestion of an asthma medication. J Am Acad Dermatol 2000;43:664-9. [Crossref] [PubMed]
- Engström K, Vahter M, Mlakar SJ, et al. Polymorphisms in arsenic(+III oxidation state) methyltransferase (AS3MT) predict gene expression of AS3MT as well as arsenic metabolism. Environ Health Perspect 2011;119:182-8. [Crossref] [PubMed]
- Pierce BL, Kibriya MG, Tong L, et al. Genome-wide association study identifies chromosome 10q24.32 variants associated with arsenic metabolism and toxicity phenotypes in Bangladesh. PLoS Genet 2012;8:e1002522. [Crossref] [PubMed]
- Srinivas N, Rachakonda S, Hielscher T, et al. Telomere length, arsenic exposure and risk of basal cell carcinoma of skin. Carcinogenesis 2019;40:715-23. [Crossref] [PubMed]
- Gailani MR, Leffell DJ, Ziegler A, et al. Relationship between sunlight exposure and a key genetic alteration in basal cell carcinoma. J Natl Cancer Inst 1996;88:349-54. [Crossref] [PubMed]
- Kim MY, Park HJ, Baek SC, et al. Mutations of the p53 and PTCH gene in basal cell carcinomas: UV mutation signature and strand bias. J Dermatol Sci 2002;29:1-9. [Crossref] [PubMed]
- Teh MT, Blaydon D, Chaplin T, et al. Genomewide single nucleotide polymorphism microarray mapping in basal cell carcinomas unveils uniparental disomy as a key somatic event. Cancer Res 2005;65:8597-603. [Crossref] [PubMed]
- Santos DC, Zaphiropoulos PG, Neto CF, et al. PTCH1 gene mutations in exon 17 and loss of heterozygosity on D9S180 microsatellite in sporadic and inherited human basal cell carcinomas. Int J Dermatol 2011;50:838-43. [Crossref] [PubMed]
- Huang YS, Bu DF, Li XY, et al. Unique features of PTCH1 mutation spectrum in Chinese sporadic basal cell carcinoma. J Eur Acad Dermatol Venereol 2013;27:235-41. [Crossref] [PubMed]
- Maturo MG, Rachakonda S, Heidenreich B, et al. Coding and noncoding somatic mutations in candidate genes in basal cell carcinoma. Sci Rep 2020;10:8005. [Crossref] [PubMed]
- Rady P, Scinicariello F, Wagner RF Jr, et al. p53 mutations in basal cell carcinomas. Cancer Res 1992;52:3804-6. [PubMed]
- Ziegler A, Leffell DJ, Kunala S, et al. Mutation hotspots due to sunlight in the p53 gene of nonmelanoma skin cancers. Proc Natl Acad Sci U S A 1993;90:4216-20. [Crossref] [PubMed]
- Rosenstein BS, Phelps RG, Weinstock MA, et al. p53 mutations in basal cell carcinomas arising in routine users of sunscreens. Photochem Photobiol 1999;70:798-806. [Crossref] [PubMed]
- Bolshakov S, Walker CM, Strom SS, et al. p53 mutations in human aggressive and nonaggressive basal and squamous cell carcinomas. Clin Cancer Res 2003;9:228-34. [PubMed]
- Wang YM, Huang YS, Ma ZH, et al. Frequency and features of TP53 mutation in 30 Chinese patients with sporadic basal cell carcinoma. Clin Exp Dermatol 2014;39:829-34. [Crossref] [PubMed]
- Iwasaki JK, Srivastava D, Moy RL, et al. The molecular genetics underlying basal cell carcinoma pathogenesis and links to targeted therapeutics. J Am Acad Dermatol 2012;66:e167-78. [Crossref] [PubMed]
- Sławińska M, Zabłotna M, Gleń J, et al. STAT3 polymorphisms and IL-6 polymorphism are associated with the risk of basal cell carcinoma in patients from northern Poland. Arch Dermatol Res 2019;311:697-704. [Crossref] [PubMed]
Cite this article as: Ju S, Fan W, Rokohl AC, Guo Y, Kakkassery V, Heindl LM. Genetic factors underlying basal cell carcinoma risk: a narrative review. Front Oral Maxillofac Med 2023;5:20.